Major Research Areas
Vector Competence for Dengue Transmission
Natural variation in dengue transmission ability across mosquito populations offers opportunities for genetic control strategies.
Key Findings:
- 60% of Ugandan mosquitoes are naturally resistant to dengue
- Brazilian populations show 32-47% susceptibility
- Distinct midgut and salivary gland infection barriers identified
Current Approach (Regents Fellowship):
Single-cell RNA-seq on phenotyped females reveals cellular mechanisms of resistance. Partnering with California MVCDs for population sampling, combining linked-read sequencing with single-cell transcriptomics.
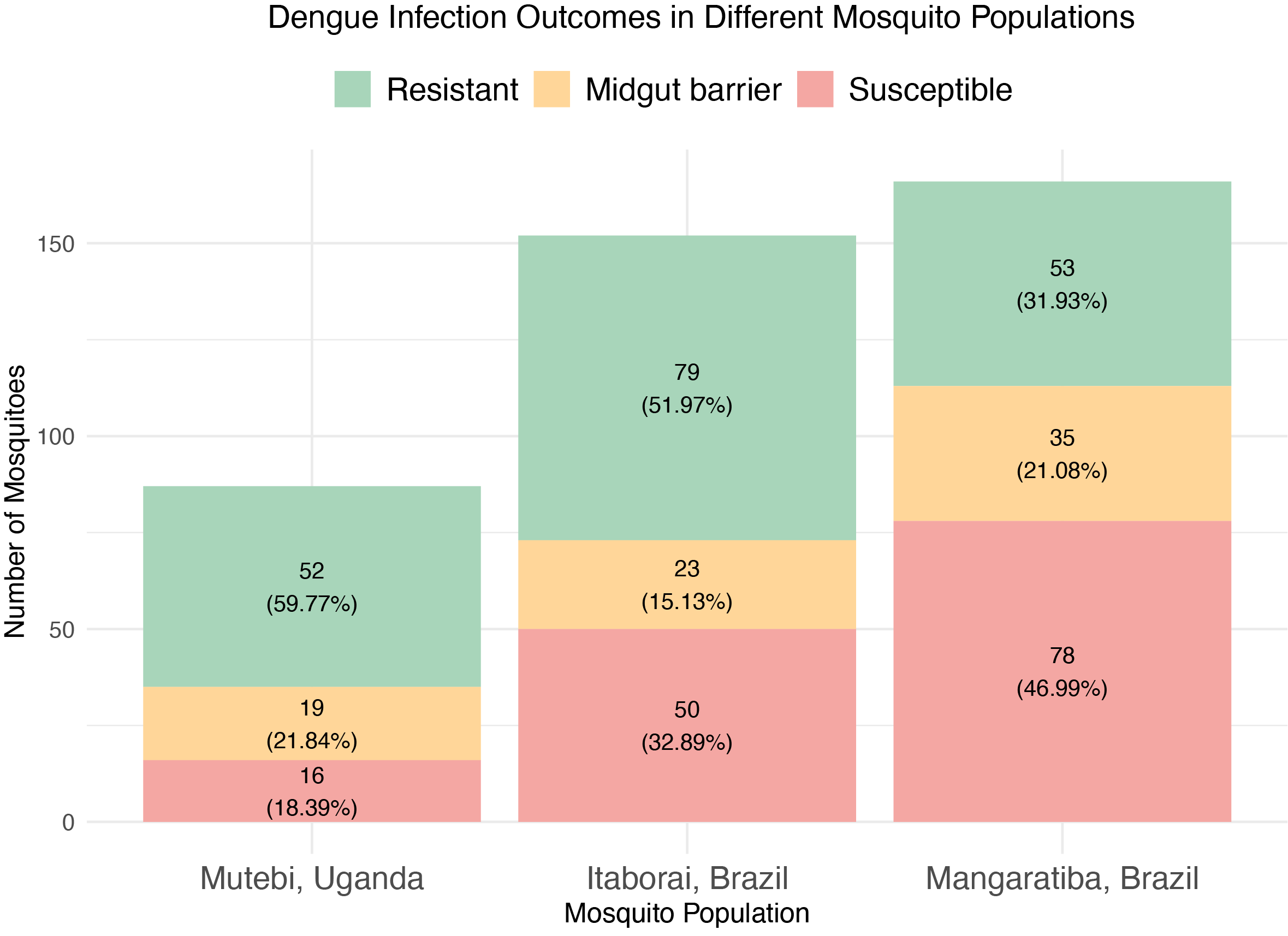
Population-level variation in dengue virus susceptibility across three Aedes aegypti populations.
Evolution of Insecticide Resistance
We study the genomic basis of insecticide resistance in Aedes aegypti populations worldwide. Our research has revealed independent origins of knockdown resistance (kdr) mutations and identified novel loci associated with pyrethroid resistance through genome-wide association studies.
Key Findings:
- Independent evolution of F1534C kdr mutation in global populations
- Discovery of new resistance-associated loci through GWAS
- Development of the Aealbo-chip for population genomics
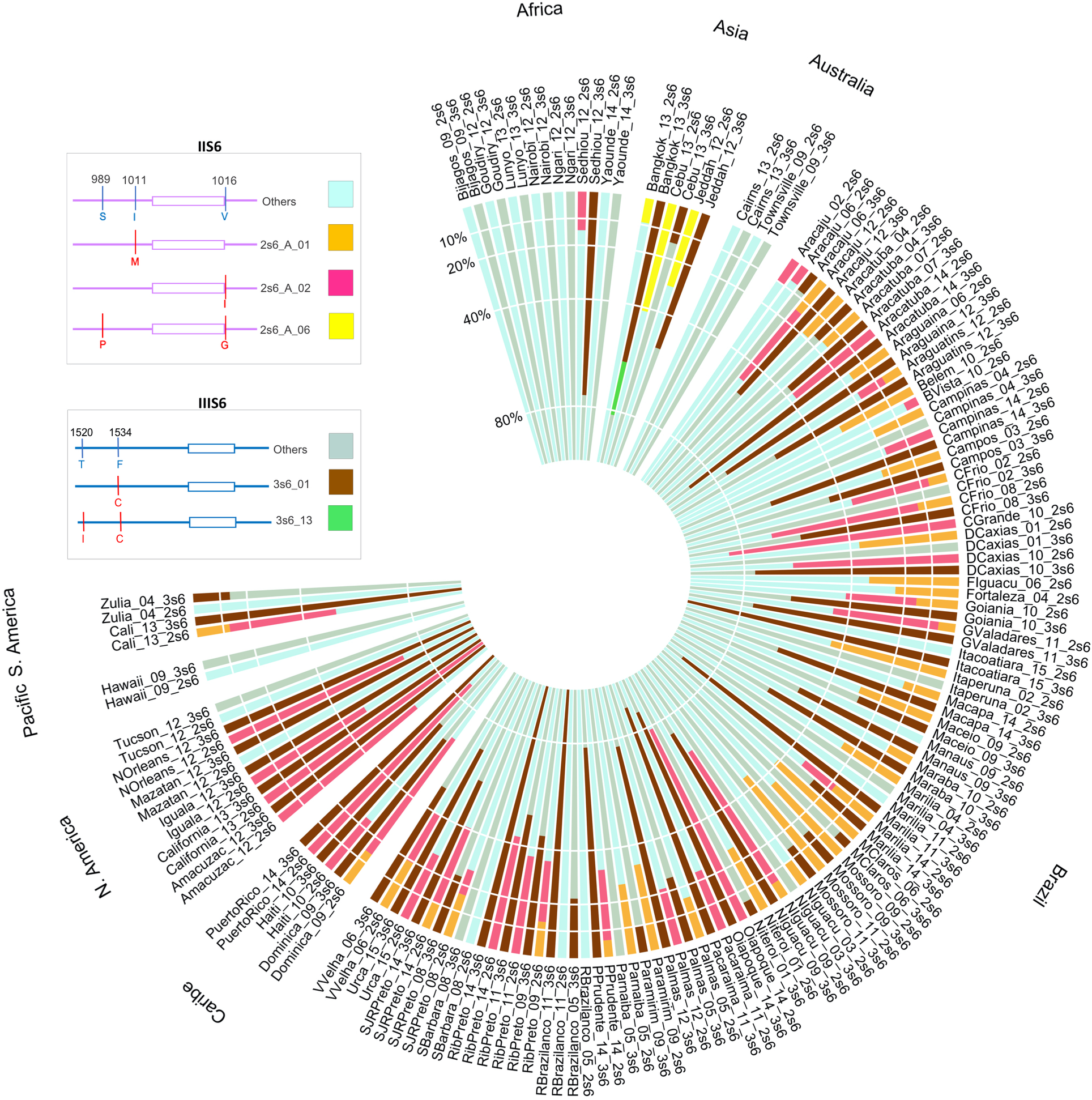
Global distribution of knockdown resistance mutations in Aedes aegypti populations. View in publication
Photoperiodic Diapause Evolution
First GWAS revealing how invasive Aedes albopictus rapidly evolved cold tolerance within ~30 years of North American invasion.
Key Findings:
- 5 SNPs on chromosome 1 control diapause entry
- Max-like protein X (MLX), part of bHLH transcription factor complexes
- Cytochrome P450s regulate metabolic shutdown
- Homeobox and Rab11 genes arrest development
Methods & Future:
Common garden GWAS with Florida lab strain under 8L:16D vs 16L:8D photoperiods. Used Aealbo-chip to overcome 70% repetitive genome. Now implementing linked-read sequencing for 3-5× power increase with wild populations from diverse climates.
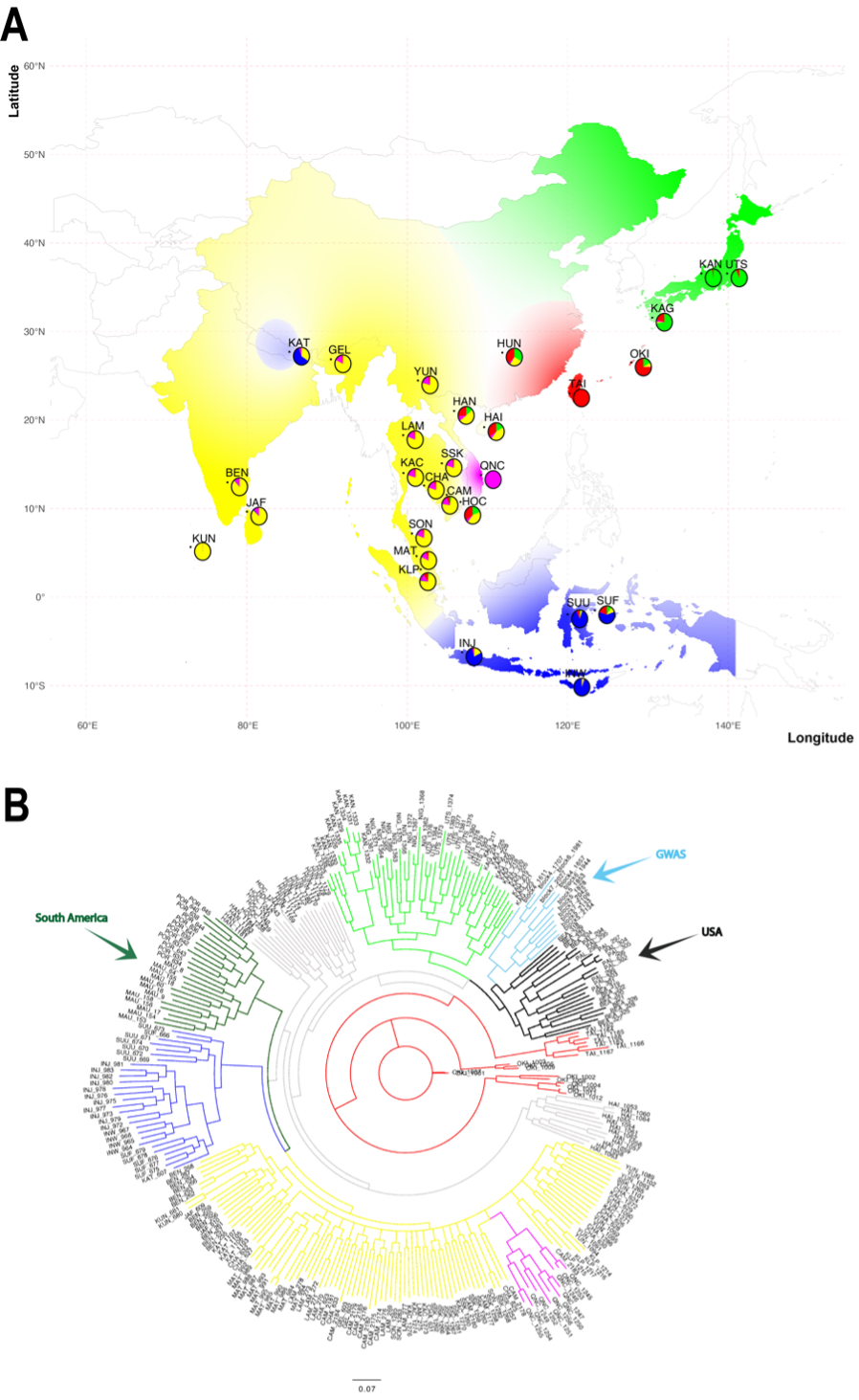
(A) Geographic distribution of genetic groups of Aedes albopictus across Asia. (B) Phylogenetic tree showing diapause and non-diapause populations
Global Population Genomics
Understanding how mosquitoes spread globally and evolve is crucial for predicting and preventing disease outbreaks. I contributed to two recent large-scale genomic studies revealing the complex evolutionary histories and invasion patterns of disease vectors worldwide.
Crawford et al. (2025) Science - Global Aedes aegypti Evolution:
A landmark 8-year study led by Dr. Jacob Crawford (Verily/Alphabet) sequenced 1,206 Aedes aegypti genomes, revealing:
- Unexpected origin: The invasive mosquito form evolved in the Americas, not Africa as long believed
- Human history shaped mosquito evolution: The Atlantic Slave Trade inadvertently created today's most efficient dengue vector
- Four evolutionary eras identified: From African forest mosquito to global urban pest
- Resistance patterns revealed: Contemporary mosquitoes returning to Africa carry insecticide resistance genes
- Ongoing admixture: Secondary contact zones in South America show active mixing between ancestral and invasive forms
Corley et al. (2025) Ecology and Evolution - European Aedes albopictus Invasion:
Dr. Margaret K. Corley from Yale University led a comprehensive study of Aedes albopictus invasion across Europe, discovering:
- Multiple invasions: At least three independent introduction events shaped European populations
- Rapid cold adaptation: Selection signatures in genes for cold tolerance and photoperiod response
- Geographic barriers: The Alps limit gene flow between Mediterranean and continental populations
- Early resistance evolution: Low-frequency kdr mutations detected in Italian and French populations
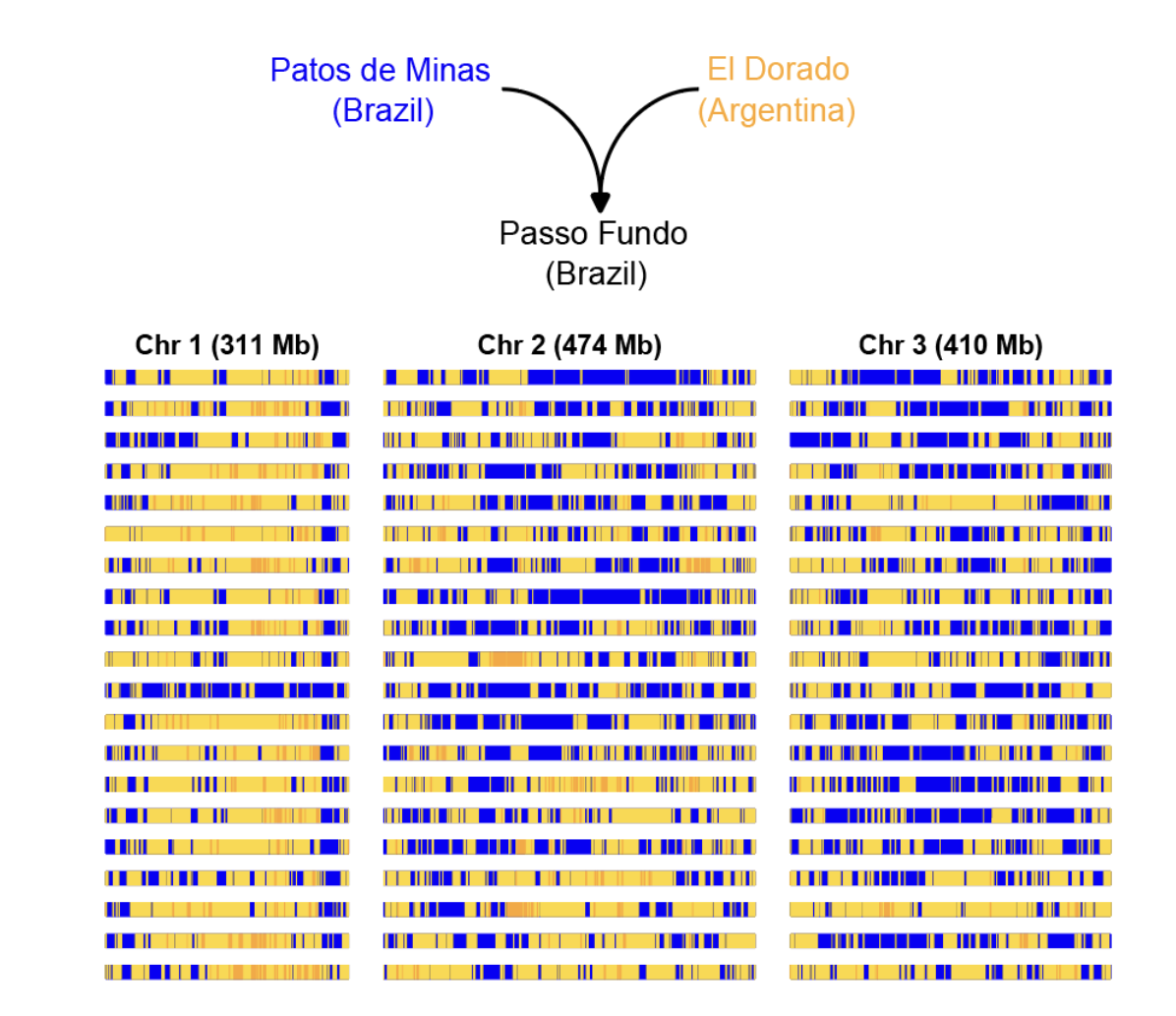
Admixture tract analysis revealing secondary contact between ancestral and invasive Aedes aegypti. View in publication
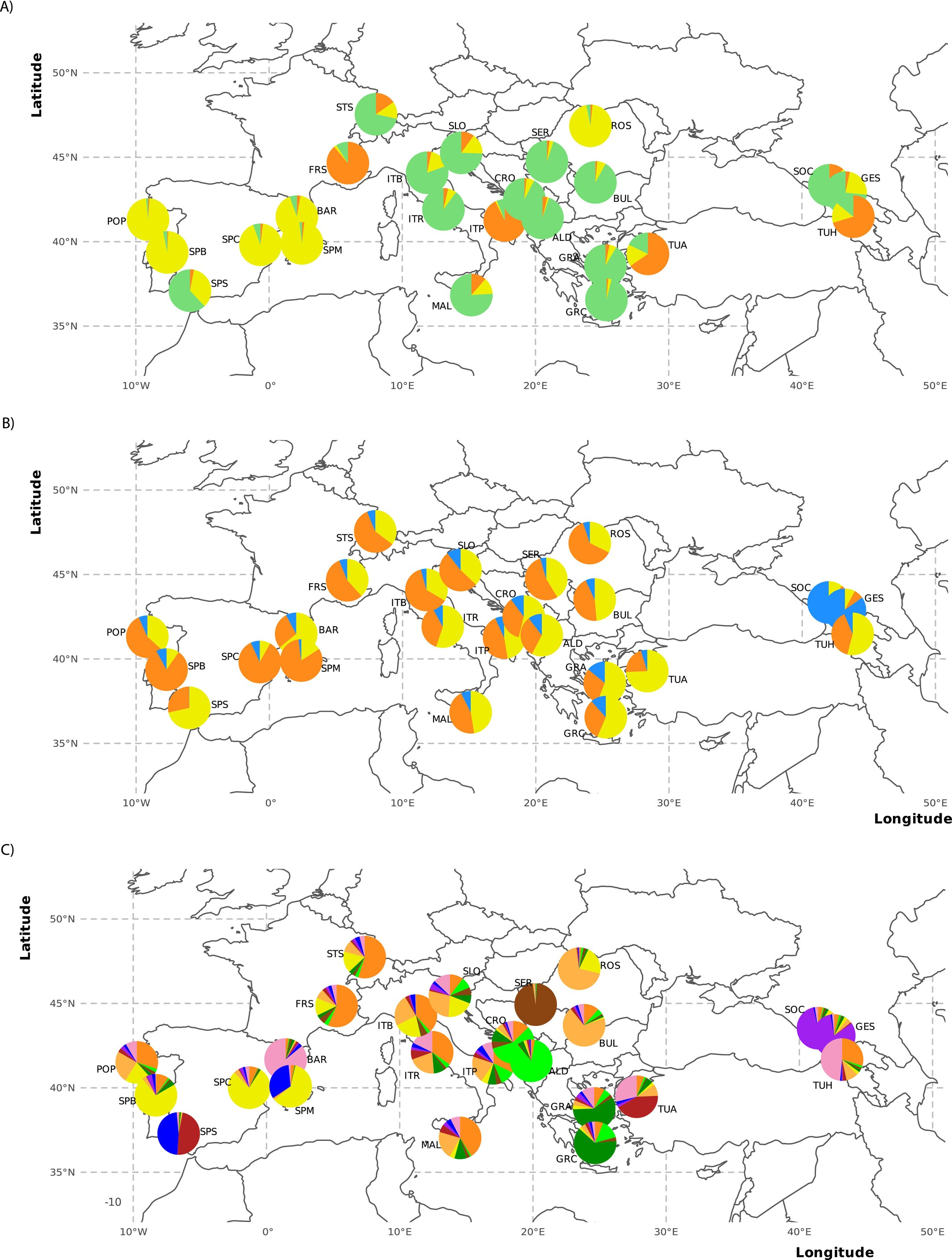
Multiple independent invasions of Aedes albopictus across Europe. View in publication







